Basics: An example workflow
Please make sure that you have activated the environment we created before, and that you have an open terminal in the working directory you have created.
A Snakemake workflow is defined by specifying rules in a Snakefile. Rules decompose the workflow into small steps (for example, the application of a single tool) by specifying how to create sets of output files from sets of input files. Snakemake automatically determines the dependencies between the rules by matching file names.
The Snakemake language extends the Python language, adding syntactic structures for rule definition and additional controls. All added syntactic structures begin with a keyword followed by a code block that is either in the same line or indented and consisting of multiple lines. The resulting syntax resembles that of original Python constructs.
In the following, we will introduce the Snakemake syntax by creating an example workflow. The workflow comes from the domain of genome analysis. It maps sequencing reads to a reference genome and calls variants on the mapped reads. The tutorial does not require you to know what this is about. Nevertheless, we provide some background in the following paragraph.
Background
The genome of a living organism encodes its hereditary information. It serves as a blueprint for proteins, which form living cells, carry information and drive chemical reactions. Differences between species, populations or individuals can be reflected by differences in the genome. Certain variants can cause syndromes or predisposition for certain diseases, or cause cancerous growth in the case of tumour cells that have accumulated changes with respect to healthy cells. This makes the genome a major target of biological and medical research. Today, it is often analyzed with DNA sequencing, producing gigabytes of data from a single biological sample (for example a biopsy of some tissue). For technical reasons, DNA sequencing cuts the DNA of a sample into millions of small pieces, called reads. In order to recover the genome of the sample, one has to map these reads against a known reference genome (for example, the human one obtained during the famous human genome project). This task is called read mapping. Often, it is of interest where an individual genome is different from the species-wide consensus represented with the reference genome. Such differences are called variants. They are responsible for harmless individual differences (like eye color), but can also cause diseases like cancer. By investigating the differences between the mapped reads and the reference sequence at a particular genome position, variants can be detected. This is a statistical challenge, because they have to be distinguished from artifacts generated by the sequencing process.
Step 1: Mapping reads
Our first Snakemake rule maps reads of a given sample to a given reference genome (see Background).
For this, we will use the tool bwa, specifically the subcommand bwa mem.
In the working directory, create a new file called Snakefile with an editor of your choice.
We propose to use the integrated development environment (IDE) tool Visual Studio Code, since it provides a good syntax highlighting Snakemake extension and a remote extension for directly using the IDE on a remote server.
In the Snakefile, define the following rule:
rule bwa_map:
input:
"data/genome.fa",
"data/samples/A.fastq"
output:
"mapped_reads/A.bam"
shell:
"bwa mem {input} | samtools view -Sb - > {output}"
A Snakemake rule has a name (here bwa_map) and a number of directives, here input, output and shell.
The input and output directives are followed by lists of files that are expected to be used or created by the rule.
In the simplest case, these are just explicit Python strings.
The shell directive is followed by a Python string containing the shell command to execute.
In the shell command string, we can refer to elements of the rule via braces notation (similar to the Python format function).
Here, we refer to the output file by specifying {output} and to the input files by specifying {input}.
Since the rule has multiple input files, Snakemake will concatenate them, separated by a whitespace.
In other words, Snakemake will replace {input} with data/genome.fa data/samples/A.fastq before executing the command.
The shell command invokes bwa mem with reference genome and reads, and pipes the output into samtools which creates a compressed BAM file containing the alignments.
The output of samtools is redirected into the output file defined by the rule with >.
When a workflow is executed, Snakemake tries to generate given target files. Target files can be specified via the command line. By executing
$ snakemake -np mapped_reads/A.bam
in the working directory containing the Snakefile, we tell Snakemake to generate the target file mapped_reads/A.bam.
Since we used the -n (or --dry-run) flag, Snakemake will only show the execution plan instead of actually performing the steps.
The -p flag instructs Snakemake to also print the resulting shell command for illustration.
To generate the target files, Snakemake applies the rules given in the Snakefile in a top-down way.
The application of a rule to generate a set of output files is called job.
For each input file of a job, Snakemake again (i.e. recursively) determines rules that can be applied to generate it.
This yields a directed acyclic graph (DAG) of jobs where the edges represent dependencies.
So far, we only have a single rule, and the DAG of jobs consists of a single node.
Nevertheless, we can execute our workflow with
$ snakemake --cores 1 mapped_reads/A.bam
Whenever executing a workflow, you need to specify the number of cores to use.
For this tutorial, we will use a single core for now.
Later you will see how parallelization works.
Note that, after completion of above command, Snakemake will not try to create mapped_reads/A.bam again, because it is already present in the file system.
Snakemake only re-runs jobs if one of the input files is newer than one of the output files or one of the input files will be updated by another job.
Step 2: Generalizing the read mapping rule
Obviously, the rule will only work for a single sample with reads in the file data/samples/A.fastq.
However, Snakemake allows generalizing rules by using named wildcards.
Simply replace the A in the second input file and in the output file with the wildcard {sample}, leading to
rule bwa_map:
input:
"data/genome.fa",
"data/samples/{sample}.fastq"
output:
"mapped_reads/{sample}.bam"
shell:
"bwa mem {input} | samtools view -Sb - > {output}"
When Snakemake determines that this rule can be applied to generate a target file by replacing the wildcard {sample} in the output file with an appropriate value, it will propagate that value to all occurrences of {sample} in the input files and thereby determine the necessary input for the resulting job.
Note that you can have multiple wildcards in your file paths, however, to avoid conflicts with other jobs of the same rule, all output files of a rule have to contain exactly the same wildcards.
When executing
$ snakemake -np mapped_reads/B.bam
Snakemake will determine that the rule bwa_map can be applied to generate the target file by replacing the wildcard {sample} with the value B.
In the output of the dry-run, you will see how the wildcard value is propagated to the input files and all filenames in the shell command.
You can also specify multiple targets, for example:
$ snakemake -np mapped_reads/A.bam mapped_reads/B.bam
Some Bash magic can make this particularly handy. For example, you can alternatively compose our multiple targets in a single pass via
$ snakemake -np mapped_reads/{A,B}.bam
Note that this is not a special Snakemake syntax.
Bash is just applying its brace expansion to the set {A,B}, creating the given path for each element and separating the resulting paths by a whitespace.
In both cases, you will see that Snakemake only proposes to create the output file mapped_reads/B.bam.
This is because you already executed the workflow before (see the previous step) and no input file is newer than the output file mapped_reads/A.bam.
You can update the file modification date of the input file
data/samples/A.fastq via
$ touch data/samples/A.fastq
and see how Snakemake wants to re-run the job to create the file mapped_reads/A.bam by executing
$ snakemake -np mapped_reads/A.bam mapped_reads/B.bam
Step 3: Sorting read alignments
For later steps, we need the read alignments in the BAM files to be sorted.
This can be achieved with the samtools sort command.
We add the following rule beneath the bwa_map rule:
rule samtools_sort:
input:
"mapped_reads/{sample}.bam"
output:
"sorted_reads/{sample}.bam"
shell:
"samtools sort -T sorted_reads/{wildcards.sample} "
"-O bam {input} > {output}"
This rule will take the input file from the mapped_reads directory and store a sorted version in the sorted_reads directory.
Note that Snakemake automatically creates missing directories before jobs are executed.
For sorting, samtools requires a prefix specified with the flag -T.
Here, we need the value of the wildcard sample.
Snakemake allows to access wildcards in the shell command via the wildcards object that has an attribute with the value for each wildcard.
When issuing
$ snakemake -np sorted_reads/B.bam
you will see how Snakemake wants to run first the rule bwa_map and then the rule samtools_sort to create the desired target file:
as mentioned before, the dependencies are resolved automatically by matching file names.
Step 4: Indexing read alignments and visualizing the DAG of jobs
Next, we need to use samtools again to index the sorted read alignments so that we can quickly access reads by the genomic location they were mapped to. This can be done with the following rule:
rule samtools_index:
input:
"sorted_reads/{sample}.bam"
output:
"sorted_reads/{sample}.bam.bai"
shell:
"samtools index {input}"
Having three steps already, it is a good time to take a closer look at the resulting directed acyclic graph (DAG) of jobs. By executing
$ snakemake --dag sorted_reads/{A,B}.bam.bai | dot -Tsvg > dag.svg
we create a visualization of the DAG using the dot command provided by Graphviz.
For the given target files, Snakemake specifies the DAG in the dot language and pipes it into the dot command, which renders the definition into SVG format.
The rendered DAG is piped into the file dag.svg and will look similar to this:
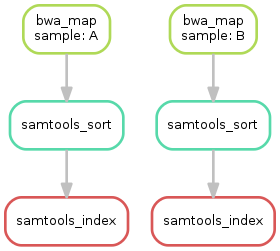
The DAG contains a node for each job with the edges connecting them representing the dependencies. The frames of jobs that don’t need to be run (because their output is up-to-date) are dashed. For rules with wildcards, the value of the wildcard for the particular job is displayed in the job node.
Exercise
Run parts of the workflow using different targets. Recreate the DAG and see how different rules’ frames become dashed because their output is present and up-to-date.
Step 5: Calling genomic variants
The next step in our workflow will aggregate the mapped reads from all samples and jointly call genomic variants on them (see Background). For the variant calling, we will combine the two utilities samtools and bcftools. Snakemake provides a helper function for collecting input files that helps us to describe the aggregation in this step. With
expand("sorted_reads/{sample}.bam", sample=SAMPLES)
we obtain a list of files where the given pattern "sorted_reads/{sample}.bam" was formatted with the values in a given list of samples SAMPLES, i.e.
["sorted_reads/A.bam", "sorted_reads/B.bam"]
The function is particularly useful when the pattern contains multiple wildcards. For example,
expand("sorted_reads/{sample}.{replicate}.bam", sample=SAMPLES, replicate=[0, 1])
would create the product of all elements of SAMPLES and the list [0, 1], yielding
["sorted_reads/A.0.bam", "sorted_reads/A.1.bam", "sorted_reads/B.0.bam", "sorted_reads/B.1.bam"]
Here, we use only the simple case of expand.
We first let Snakemake know which samples we want to consider.
Remember that Snakemake works backwards from requested output, and not from available input.
Thus, it does not automatically infer all possible output from, for example, the fastq files in the data folder.
Also remember that Snakefiles are in principle Python code enhanced by some declarative statements to define workflows.
Hence, we can define the list of samples ad-hoc in plain Python at the top of the Snakefile:
SAMPLES = ["A", "B"]
Later, we will learn about more sophisticated ways like config files. But for now, this is enough so that we can add the following rule to our Snakefile:
rule bcftools_call:
input:
fa="data/genome.fa",
bam=expand("sorted_reads/{sample}.bam", sample=SAMPLES),
bai=expand("sorted_reads/{sample}.bam.bai", sample=SAMPLES)
output:
"calls/all.vcf"
shell:
"bcftools mpileup -f {input.fa} {input.bam} | "
"bcftools call -mv - > {output}"
With multiple input or output files, it is sometimes handy to refer to them separately in the shell command.
This can be done by specifying names for input or output files, for example with fa=....
The files can then be referred to in the shell command by name, for example with {input.fa}.
For long shell commands like this one, it is advisable to split the string over multiple indented lines.
Python will automatically merge it into one.
Further, you will notice that the input or output file lists can contain arbitrary Python statements, as long as it returns a string, or a list of strings.
Here, we invoke our expand function to aggregate over the aligned reads of all samples.
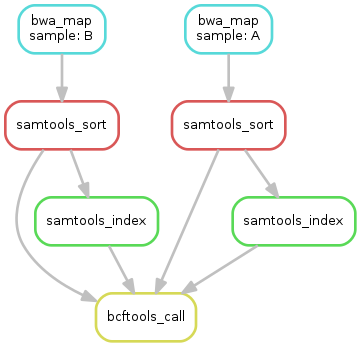
Exercise
obtain the updated DAG of jobs for the target file
calls/all.vcf, it should look like this:
Step 6: Using custom scripts
Usually, a workflow not only consists of invoking various tools, but also contains custom code to for example calculate summary statistics or create plots.
While Snakemake also allows you to directly write Python code inside a rule, it is usually reasonable to move such logic into separate scripts.
For this purpose, Snakemake offers the script directive.
Add the following rule to your Snakefile:
rule plot_quals:
input:
"calls/all.vcf"
output:
"plots/quals.svg"
script:
"scripts/plot-quals.py"
With this rule, we will eventually generate a histogram of the quality scores that have been assigned to the variant calls in the file calls/all.vcf.
The actual Python code to generate the plot is hidden in the script scripts/plot-quals.py.
Script paths are always relative to the referring Snakefile.
In the script, all properties of the rule like input, output, wildcards, etc. are available as attributes of a global snakemake object.
Create the file scripts/plot-quals.py, with the following content:
import matplotlib
matplotlib.use("Agg")
import matplotlib.pyplot as plt
from pysam import VariantFile
quals = [record.qual for record in VariantFile(snakemake.input[0])]
plt.hist(quals)
plt.savefig(snakemake.output[0])
Although there are other strategies to invoke separate scripts from your workflow (for example, invoking them via shell commands), the benefit of this is obvious: the script logic is separated from the workflow logic (and can even be shared between workflows), but boilerplate code like the parsing of command line arguments is unnecessary.
Apart from Python scripts, it is also possible to use R scripts. In R scripts,
an S4 object named snakemake analogous to the Python case above is available and
allows access to input and output files and other parameters. Here, the syntax
follows that of S4 classes with attributes that are R lists, for example we can access
the first input file with snakemake@input[[1]] (note that the first file does
not have index 0 here, because R starts counting from 1). Named input and output
files can be accessed in the same way, by just providing the name instead of an
index, for example snakemake@input[["myfile"]].
For details and examples, see the External scripts section in the Documentation.
Step 7: Adding a target rule
So far, we always executed the workflow by specifying a target file at the command line.
Apart from filenames, Snakemake also accepts rule names as targets if the requested rule does not have wildcards.
Hence, it is possible to write target rules collecting particular subsets of the desired results or all results.
Moreover, if no target is given at the command line, Snakemake will define the first rule of the Snakefile as the target.
Hence, it is best practice to have a rule all at the top of the workflow which has all typically desired target files as input files.
Here, this means that we add a rule
rule all:
input:
"plots/quals.svg"
to the top of our workflow. When executing Snakemake with
$ snakemake -n
the execution plan for creating the file plots/quals.svg, which contains and summarizes all our results, will be shown.
Note that, apart from Snakemake considering the first rule of the workflow as the default target, the order of rules in the Snakefile is arbitrary and does not influence the DAG of jobs.
Exercise
Create the DAG of jobs for the complete workflow.
Execute the complete workflow and have a look at the resulting
plots/quals.svg.Snakemake provides handy flags for forcing re-execution of parts of the workflow. Have a look at the command line help with
snakemake --helpand search for the flag--forcerun. Then, use this flag to re-execute the rulesamtools_sortand see what happens.Snakemake displays the reason for each job (under
reason:). Perform a dry-run that forces some rules to be reexecuted (using the--forcerunflag in combination with some rulename) to understand the decisions of Snakemake.
Summary
In total, the resulting workflow looks like this:
SAMPLES = ["A", "B"]
rule all:
input:
"plots/quals.svg"
rule bwa_map:
input:
"data/genome.fa",
"data/samples/{sample}.fastq"
output:
"mapped_reads/{sample}.bam"
shell:
"bwa mem {input} | samtools view -Sb - > {output}"
rule samtools_sort:
input:
"mapped_reads/{sample}.bam"
output:
"sorted_reads/{sample}.bam"
shell:
"samtools sort -T sorted_reads/{wildcards.sample} "
"-O bam {input} > {output}"
rule samtools_index:
input:
"sorted_reads/{sample}.bam"
output:
"sorted_reads/{sample}.bam.bai"
shell:
"samtools index {input}"
rule bcftools_call:
input:
fa="data/genome.fa",
bam=expand("sorted_reads/{sample}.bam", sample=SAMPLES),
bai=expand("sorted_reads/{sample}.bam.bai", sample=SAMPLES)
output:
"calls/all.vcf"
shell:
"bcftools mpileup -f {input.fa} {input.bam} | "
"bcftools call -mv - > {output}"
rule plot_quals:
input:
"calls/all.vcf"
output:
"plots/quals.svg"
script:
"scripts/plot-quals.py"